Over the past few weeks the Science Practice studio has been buzzing with talk of Brexit. As with much of the UK, and seemingly both sides of the campaign, we were taken by surprise by the decision of 17,410,742 people to leave the European Union. Probably unsurprisingly for a company with 5 nationalities represented in a workforce of 7 we were unanimously in favour of remaining within the EU. In fact we fit almost all stereotypes of a Remain voter - multi-national, under 35, based in London and working in the science sector. It is fair to say the mood in the office on the 24th of June was pretty grim…
Over the years Science Practice has worked with many scientists and organisations across the EU, including the Commission itself. We have directly benefited from the willingness to collaborate across borders that the EU holds as a core ideology. Our most obvious example is a project in which we helped design challenge prizes for the European commission, involving 68 interviews with EU experts, and a year long collaboration with the University Scuola Superiore Sant’Anna in Italy.
At the time of writing the date for the UK to begin the process of leaving the EU appears to be shifting ever further away. Still, with our new prime minister Theresa May stating that “Brexit means Brexit” we felt it was important to engage with the conversation on Brexit and science. What did the membership of the EU mean for science? What could we lose? And what we should fight for?
What we had.
The majority consensus is that UK science got a pretty good deal from the EU. As a sector it in fact had a net gain in terms of funding, receiving around £1.2 billion a year for research projects, a figure that makes up nearly a fifth of all UK funding from the EU. In the latest funding programme - Horizon 2020 - the UK has received 15.4% of funds allocated so far, second only to Germany. As a whole, the EU is the world leader in terms of scientific output. With 22% of all scientists in the world, the EU puts out over a third of the world’s scientific articles each year.
The UK has a lot of EU nationals working in its academic institutions. Freedom of movement, combined with the very high reputation of British universities made the UK very attractive for talented researchers across the EU. 15% of all researchers in UK institutions are EU nationals, with this number rising to closer to 20% in the top-ranking universities.
This pan-european collaboration is widely seen as positive amongst EU scientists. Just under a third of all EU academics have worked in a different EU country, with 80% of them saying that this experience has had a positive impact on their research skills.
Outside of the strictly quantifiable, the UK had a strong influence in the direction of EU science policy. Many advisory positions contain or contained UK scientists, such as the (admittedly short-lived) former Chief Scientific advisor to the Commission - Prof Anne Glover, and Prof Julia Slingo who is a member of the replacement science advisory mechanism.
One aspect of EU membership that is sometimes overlooked is the number of students that move to the UK to access higher education institutions. 47,000 EU students come to study in the UK each year, roughly a quarter of a total 192,000 students. It would be difficult for even the most anti-immigration campaigners to be against this yearly input of students. In 2012 they contributed £3.7 billion to the UK economy, and supported 34,000 jobs.
What now?
The overwhelming response amongst scientists and those involved in science policy is to begin making arguments to preserve as much of the current arrangements as possible. Assurances are being sought from UK science minister Jo Johnson (who has maintained his position after the cabinet reshuffle) that the UK will continue to have access to both people and money from the EU. A member of the remain campaign himself, Johnson’s initial responses have been reassuring in intent, although his ability to make a special case for science in the chaos of negotiations remains to be seen.
There are signs that the current environment of uncertainty is already causing problems for UK scientists. Reports have begun to emerge of EU research partners dropping out of proposals for Horizon 2020 projects. The House of Commons Science and Technology Committee have begun collecting stories from UK scientists to gather a picture of effects on the ground for researchers.
A particular sticking point for UK science is the seemingly strong support for a more restrictive immigration policy. Restrictions on immigration could be problematic for science based institutions in attracting and maintaining their workforces. Although it is likely that the majority of researchers and scientists would meet any new visa skills requirements, the added administrative burden, and perception of a hostile environment would most probably reduce numbers.
A less immediately obvious issue with restricting movement is access to EU funding. In 2014 Switzerland held a referendum on the freedom of movement of EU citizens. The result, which was to introduce stronger immigration controls, led to the almost immediate removal of Swiss institutions abilities to apply for Horizon 2020 funding. Since 2014 the EU’s position has softened a little, Swiss academics can now apply for around 30% of funds, but are treated as a ‘third country’ with respect to 70%. Switzerland was never an EU member state, but was treated very harshly by the EU for challenging one of its core tenets. The UK should be prepared for equally harsh treatment.
Although access to Horizon 2020 funding could be restricted, it may be possible for the UK to up spending on science through money previously allocated for EU membership. Although many sectors will be looking for this money to replace funding and subsidies that are lost, it was considered a possibility in responses to the House of Lords Science and Technology Select Committee’s investigation into EU Membership and UK Science. My personal feeling is that the UK is unlikely to match the levels of funding previously received from the EU. In times of austerity the idea that a conservative government will increase public spending on science by £1 billion a year seems far-fetched.
One area in which it is less obvious what may happen are in the regulations on science and scientific industries. For example the EU is strict on using genetic modification for food crops. Without the requirement to meet EU regulations, UK scientists and farmers may be permitted to grow GM crops more widely. There is significant support within the scientific community for greater usage of GM technologies for foods, however changing policy to be more permissive would be a controversial move.
Moving forwards
Much has been made of the attempts from supporters of Remain to push for Brexit negotiations to allow for the UK to have access to as much of the EU as possible. In reality it seems doubtful that the EU will allow this easily, early signs suggest that this would seen as encouragement of copycat ‘Brexits’ (Czech-out, Quitaly, Portugo……). For science though, it really makes sense to try and keep things as they were. Jo Johnson, Greg Clarke and Justine Greening (Secretary of Business, Energy and Industrial Strategy and Education respectively) should make a strong case for access to the European funding, and talent. The example of Switzerland should be highlighted, with the need of increased funding from within the UK emphasised if strong restrictions on immigration seem inevitable.
As Science Practice we are unlikely to be directly affected by Brexit, no contracts have been lost and our offices won’t move to Dublin, Paris or Berlin. However our ability to work across borders and engage EU researchers and academics is at risk, although the full extent of this is yet to be seen.
UK scientists are often vocal about how they hope that their sector behaves and is structured, with strong campaigns for open access and increased participation in science from women and minorities, not to mention the case for remaining in the EU. Now another cause must be added - maintained openness and collaboration outside of the UK’s borders. EU membership is not the only thing that makes UK science great, however now more than ever science needs to show that it knows no borders.
For the last 6 months we have been working with Genomics PLC to help design and prototype a new product. Genomics PLC are a very talented team in Oxford who are building applications that uncover links between genetic variation and human disease.
It has been fascinating to further develop our interest in genomics and its relationship to real-world healthcare. Although we aren’t able to speak about the details, we can say that we have learnt a lot about the potential for software to help make sense of genetic data.
From a methodological point of view, we are always striving for fruitful collaborations between the disciplines. Working with Genomics PLC provided a fantastic opportunity to get scientists, software engineers and designers working together in a productive and satisfying way.
So why is a designer interested in learning about bioinformatics?
I’ve been asked this question many times during my stay at the Wellcome Trust Genome Campus in Hinxton, where I attended the EBI Bioinformatics Summer School two weeks ago.
At Science Practice we often work with bioinformaticians helping them build expert tools. Not long ago we collaborated with the Goldman Group at the EBI and developed Sequence Bundles. Most recently we have been testing the MinION, sequencing soil metagenomes, and working with biomedical startups and companies as part of Ctrl Group. Each project requires us to sit down for a few hours or days and learn the science behind it. This learning curve is something I personally really enjoy. And because every single project we work on at Science Practice is very different from another, there is always a lot of diverse learning going on.
So my answer to the question I was asked was:
Because I’ve got a general second-hand understanding of bioinformatics, but no direct hands-on experience of how it’s done.
And we want that first-hand “done-it-myself” experience. This keeps us very close to what actually happens in the lab, in the clinic, in the field — in the very places where this insider expertise matters. That’s why I decided to spend a week in Hinxton together with 30 researchers, postdocs, and PhD candidates from around Europe to learn the science of bioinformatics in practice.

Hinxton Hall at the Wellcome Genome Campus
The course took place at the Wellcome Genome Campus which was one of the key centres involved in the seminal Human Genome Project. One of the institutes based at the Campus is EMBL European Bioinformatics Institute — the organiser of the Bioinformatics Summer School — and one of the world’s main centres for storing, curating and providing biological sequence data. The campus itself is set in rural England in vicinity of the old Hinxton Hall.
The course had a balanced mix of theory and practice. On day one Bill Pearson from the University of Virginia kicked off with a general primer in bioinformatics as an experimental science: How do we get answers to the questions that we want answered? Which questions cannot be answered by bioinformatics at all and which ones cannot be answered just yet? What are the positive and negative controls in bioinformatics? Why do we search for similarity and what are the statistics of excess similarity? And why statistically significant structure similarity can be present even in absence of statistically significant sequence similarity?
This was followed by several short tutorials on using bioinformatics resources offered by the EBI, including using the Ensembl genome browser, querying nucleotide and protein databases online, and also requesting jobs programmatically via the command line.
All that theory was an indispensable primer for launching the main activity of the Summer School on day two — a practical bioinformatics project. There were four project teams to select from:
- Genome Assembly and Annotation
- Phylogenetics
- RNA-Seq and Networks
- Structural Biology and Functional Prediction
I chose to do the Genome Assembly and Annotation project, as it had most to do with clinical genomics and genomic medicine which is in one of our areas of interest at Science Practice.
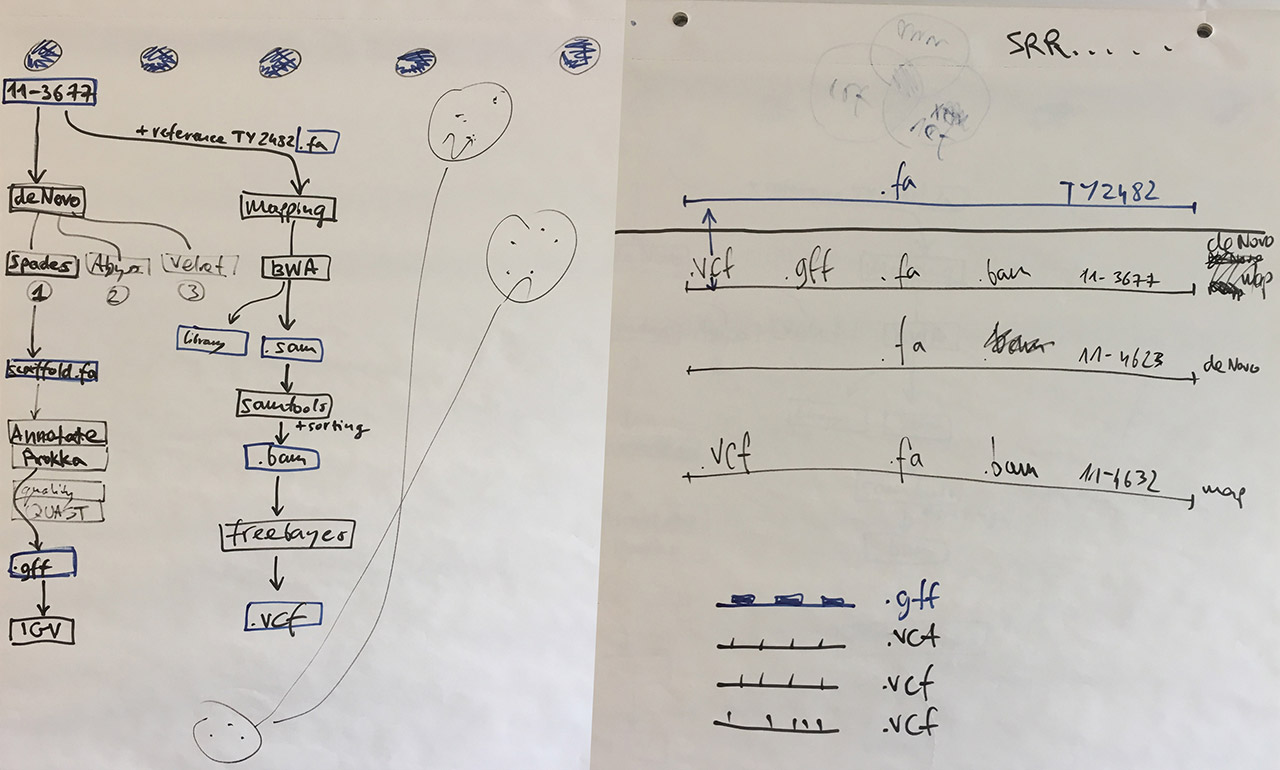
Visual notes from the bioinformatics lab
The canvas for the bioinformatics exercise was the 2011 E coli outbreak in Germany and France. The outbreak was caused by the highly pathogenic Shiga toxin-producing O104:H4 strain of Escherichia coli. Infected patients were at risk of suffering from bloody diarrhea (around 4,000 cases) which could progress to life-threatening hemolytic uremic syndrome, known as HUS (859 cases). 50 patients with severe cases of HUS died. The 2011 E coli outbreak was unusual for the rapid response in which multiplatform sequencing was brought to understand the epidemic, which is outlined in this PNAS paper.
Our task was to re-trace the investigation done during the outbreak. Working in a group of four (myself, Tracy Munthali Lunde, Massimiliano Volpe, and Dag Harald Skutlaberg) we followed these steps:
-
Obtain raw genomes from the outbreak strain sequencing experiments from the European Nucleotide Archive (data quality checked in FastQC);
-
Assemble genomes using the BWA mapping assembler — we worked with strains 11-3677, 11-4623, and 11-4632 described in the PNAS paper, and the E. coli TY-2482 reference obtained from the GigaDB database;
-
Convert, sort, index — use samtools to convert and sort assembled SAM files into an indexed binary BAM file that can be displayed in the IGV genome browser, and which can later be used for calling variants;
-
Call variants between the TY-2482 reference sequence and the assembled genomes 11-3677, 11-4623, and 11-4632 using freebayes;
-
Annotate genome using Prokka to identify which regions of the reference sequence are protein coding — this will help to understand which variants are important;
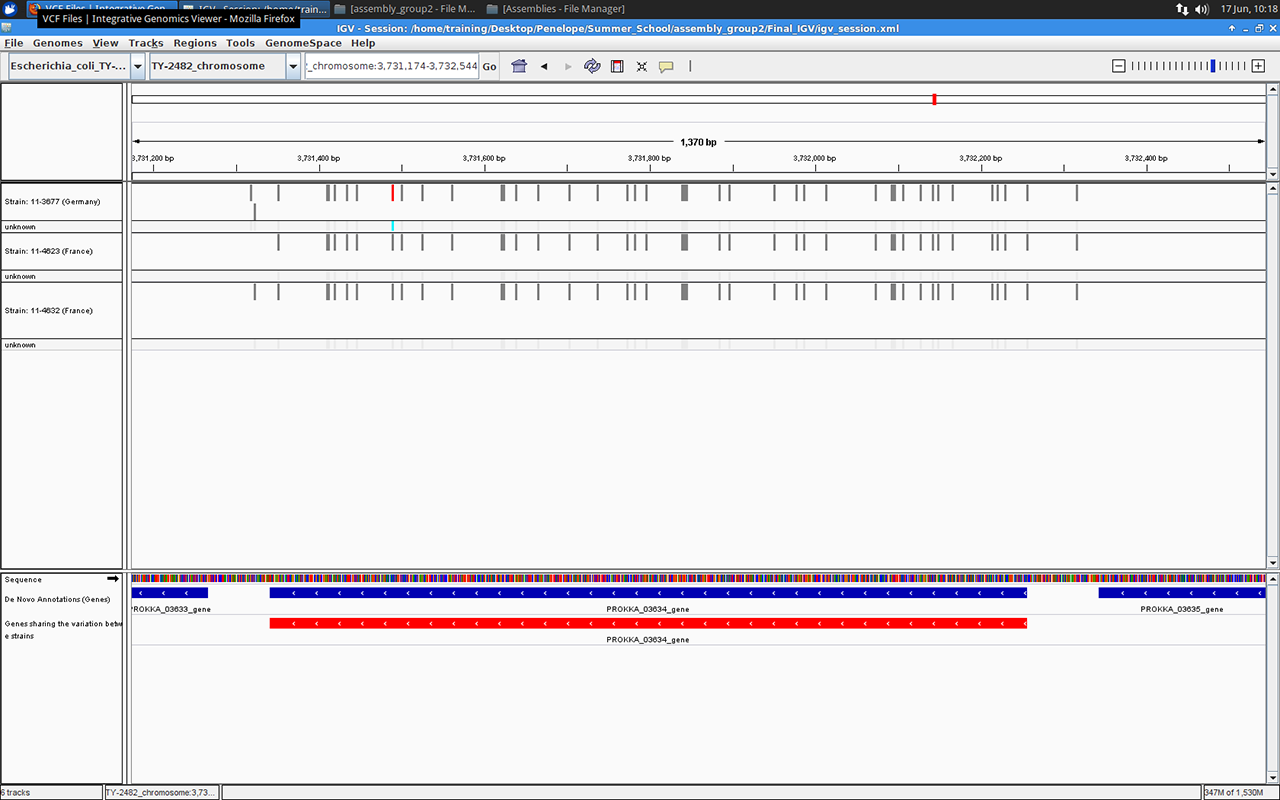
Gene number 03634 in IGV browser, screenshot by Massimiliano Volpe
We ended our project with a little investigation into the variants identified in the assembled genomes. For example we found out that about 60% of all variants in all three genomes fell within the annotated genes.
As a side project, we also managed to compare the performance of three de novo assemblers: Velvet, Abyss, and Spades. With the genome data we used, the comparison was won by Spades.
In the end, our process involved running the sequence data through a series of tools operated from the UNIX command line. Even though very simple, lacking any investigation into detailed algorithm settings and tested only on three datasets; by the end of the EBI Summer School we have succeeded to prototype our first bioinformatics pipeline.
Producing one's first ever VCF file feels great! 543 variants in a 5mbp E. coli genome. Highly toxic serotype! pic.twitter.com/dBsDNELuHQ
— Science Practice (@sciencepractice) June 15, 2016
The EBI Bioinformatics Summer School was a fantastic learning experience that will help us do even more in the prospective bioinformatics and genomics projects at Science Practice. For those interested in trying out some bioinformatics on their own, here are the Lab Notes (shared under CC-BY-4.0) that I prepared during the course. Happy assembling!
We’re looking for someone passionate about science, tech and innovation to join our team over the summer!
- Location: 83-85 Paul Street, London EC2A 4NQ, UK
- Term: 3 months
- Hours: Full-time
- Starting date: June 2016
- Salary: The internship will be paid, the rate will be negotiated with the successful applicant based on experience.
Things have been pretty lively in the studio lately and we’re looking for another pair of hands to help out with some of the research work!


By means of introduction - we’re Science Practice, a design and research company based in London. We work across a variety of areas and collaborate with scientists to help put their research into practice. Whether we’re designing challenge prizes or humanitarian ones, trying to sequence a virus without a lab or creating new methods for visualising genetic data, we’re always looking at ways to integrate design principles with the processes and methodologies of science.
The role
We’re looking to recruit a recent graduate to support our work on researching and designing challenge prizes. The role will involve:
- Supporting the team research technological innovations and their impact on addressing social problems;
- Writing key research findings and helping prepare external project documentation;
- Supporting expert engagement work - including logistical tasks such as identifying key experts in a specific field and arranging interviews with them, as well as taking part in the interviews and synthesising the main ideas;
- Working closely with a team of international partners in designing the structure of several challenge prizes;
- Completing important and often time sensitive ad hoc project tasks.
Our intern
- Passionate about technology and its potential for social impact;
- Interested in innovation and tools for supporting innovation such as challenge prizes or competitions;
- Strong academic credentials - recent graduate of a Bachelors or Masters (preferably in an engineering or technology related degree), with a First or 2.1;
- Strong research skills - ability to understand and synthesise complex information quickly;
- Excellent written and verbal communication skills;
- Highly organised, with a close attention to detail and high standards of accuracy;
- Efficient, self-motivated and able to manage priorities to meet deadlines;
- Able to work on location in London, UK.
If you’re keen and ready to give a helping hand, please send an email with your CV and a brief cover letter to Ana at af@science-practice.com. Thanks and looking forward to hearing from you soon!
The potential for Oxford Nanopore’s MinION to revolutionise genomics has been well documented, and for good reason. As we and others have mentioned before, they have drastically reduced the size of sequencers so that their device will fit in a pocket. Perhaps as importantly, the protocol to prepare samples for sequencing is significantly simpler than traditional sequencing, raising the possibility of this technology being used by more and more people interested in genetics - including us!

Our MinION running
Having recently conducted a burn-in experiment to check that our MinION works and to familiarise ourselves with the protocol, we had been racking our brains for a good experiment to use our remaining 2 flow cells for.
Sequencing for soil health
As one of our current focuses is soil health we thought it would be interesting to investigate whether we could extract DNA from the organisms found in two different soil samples, sequence them using MinION, and use the species identified to infer information about the soil properties. After some further digging on Google we were unable to find evidence of anyone else who had tried this, giving us even more reason to give it a go!
The analysis we were intending to do was made possible thanks to an app for Metrichor (Oxford Nanopore’s analysis software) called What’s In My Pot (WIMP). WIMP connects to the NCBI database of bacterial, fungal and viral genomes and matches reads sequenced by MinION to their respective species.


Mile End Park and a Bloomsbury vegetable patch were our chosen places to take soil samples
Our first steps were to identify two places to collect soil samples from, and to find a way to extract DNA from the bacteria and fungi found within them (known as the metagenome). For the soil we settled on two samples from London: one from a worn patch of ground in Mile End Park, and one from a shared vegetable patch. Our hypothesis was that there may be detectable differences between the cultivated and non-cultivated soils.
Extracting DNA from soil
DNA extraction was easier than you may expect. We were able to order a DNA extraction kit from MO BIO called Powersoil which was simple to use and took about two hours to go from sample to DNA in a lab at Imperial College London. Ordinarily a biomedical lab, we raised a few eyebrows with our soil experiments!

Our soil samples ready for DNA extraction
The next step was to find somewhere to prepare the DNA and to run the MinION sequencing. As we were well aware from our burn-in experiment, there are a number of pieces of lab equipment that are necessary that we do not have access to, and so we needed to find someone with lab space who would be willing to work with us on this mini-project.
Visiting York
Through twitter we managed to find James Chong at The University of York, who had recently given a talk on using MinION to sequence bacterial 16s ribosomal RNA, and asked if he might be interested. Thankfully for us, James was incredibly generous with his time and knowledge, introduced us to his equally helpful post-doc Anna Alessi who had been using MinION, and offered to have us visit his lab and run the experiment.
Excited to be in @insanity_one lab today @yorkuniversity! sequencing soil creatures with the #minion @nanopore pic.twitter.com/3KbyjYRoVR
— Science Practice (@sciencepractice) 5 May 2016
When we arrived in York we were warmly welcomed by James, Anna and Farah Shahi who is a medical doctor at the start of a research project using MinION and is learning lab techniques. After picking James’s brain for advice for our soil health projects we quickly got going on the MinION protocol, starting by checking whether we had enough DNA using Qubit, a fluorometer.
We were relieved to find that we had more than enough DNA, and so proceeded with the same protocol as we did for the burn-in, only this time we prepared two samples in parallel: the sample from the park and the vegetable patch. By the end of the protocol we were pleased to have two good DNA libraries that met Oxford Nanopore’s suggested quantity requirements.
Relieved to find that we extracted enough #DNA from soil to sequence on the @nanopore #minion today #dnaparty pic.twitter.com/ZnMWaqXI0G
— Science Practice (@sciencepractice) 5 May 2016
While the wet-lab component of the day went without problem, we did have some issues with running MinION itself. Firstly we were unable to get our laptop to recognise the MinION. We were aware that this could be temperamental from our experiences in the burn-in, however for this experiment it simply did not manage to acknowledge the device. Fortunately for us, James has five MinIONs permanently connected to five laptops and so we were able to use his.

Using DNA binding magnetic beads and a magnetic tube rack to isolate our DNA samples
Silent flow cells
Having decided to use James’s MinION we then tested out our two flow cells. Although we had stored them how MinION recommended, and both were visually in good condition, the test run indicated that we had 0 working pores on either flow cell. Despite consulting Anna, a bioinformatician and even the Head of Genomics, we were not able to get to the bottom of this. We suspect there was a serious underlying problem with these flow cells, rather than a degradation of pores.

A MinION flowcell, the bubbles that are visible can hinder the experiment
Fortunately Anna and James came to the rescue again, and allowed us to use two of their spare flow cells. Although they were not in excellent condition (there were visible bubbles covering the pores), and only had around 150-400 active pores, after getting this far with our experiment it would have been a shame not to finish.


At work in the lab setting up our MinION runs
We ran the MinION overnight, and in the morning, reloaded with more of the libraries we had prepared and ran again (this is a trick that James and Anna have found improves the number of usable reads). As expected the data we got was relatively poor quality, with the majority of reads not meeting Metrichor’s quality control.
Genetic sequencer may be pocket-sized but we have the entire teaching lab to ourselves to run it @nanopore #minion pic.twitter.com/q0oQEAlb4u
— Science Practice (@sciencepractice) 6 May 2016
What’s in my pot?
Despite having few good quality reads we decided to run WIMP for both samples, to see what it was able to identify in each sample, and to get a better idea of how the app works. Although we can’t draw any confident conclusions from what we found, there were some small hints that it would be possible to use MinION combined with WIMP to gather useful information about soil.
For example several bacteria capable of metabolising common environmental pollutants were identified in the sample of soil from the park (Nocardioides, V. paradoxus), while the soil from the vegetable patch, which has had manure and compost added to it, had bacteria often found in sewer sludge and wet soil (Thauera). Both samples had many different bacteria involved in the nitrogen cycle.
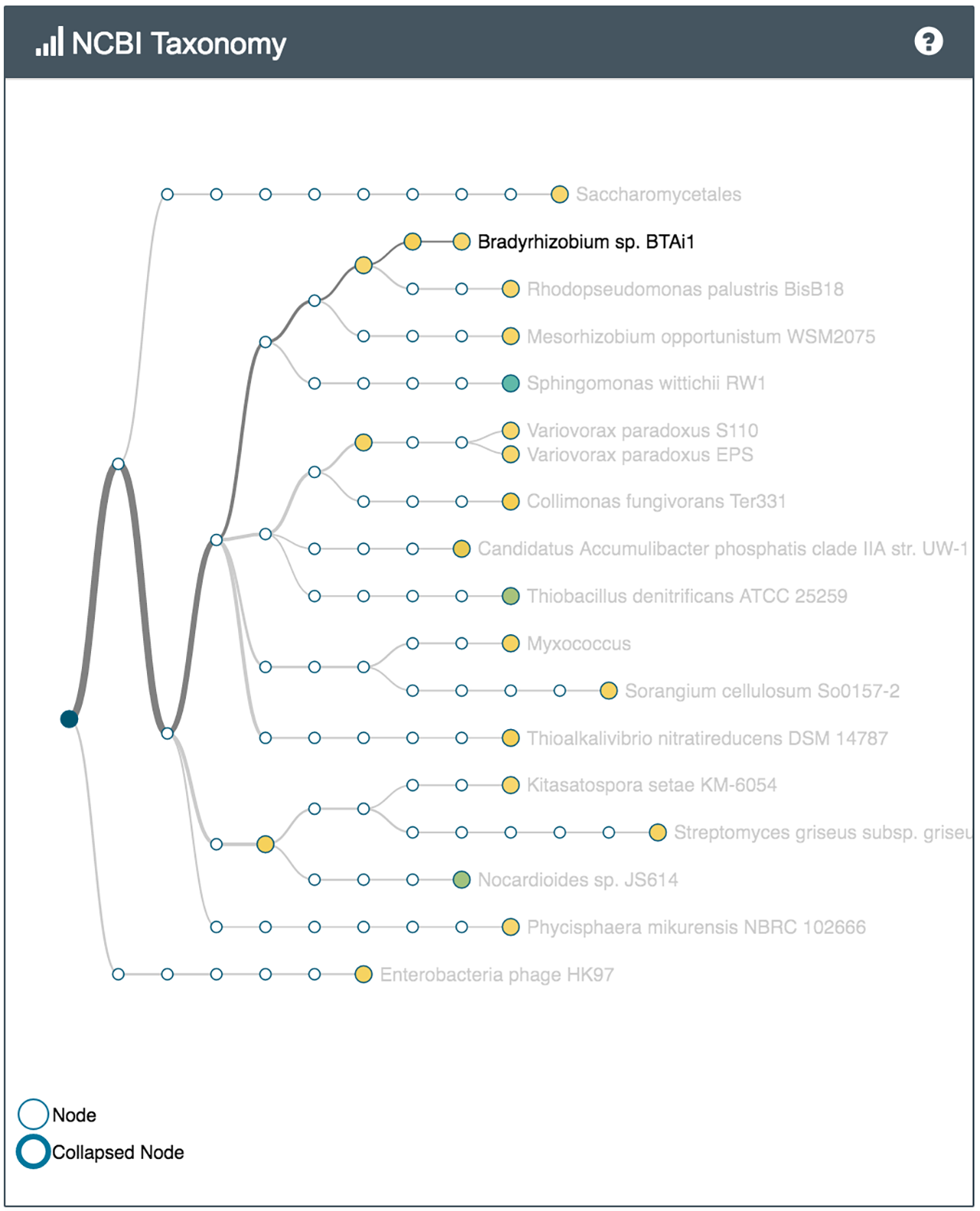
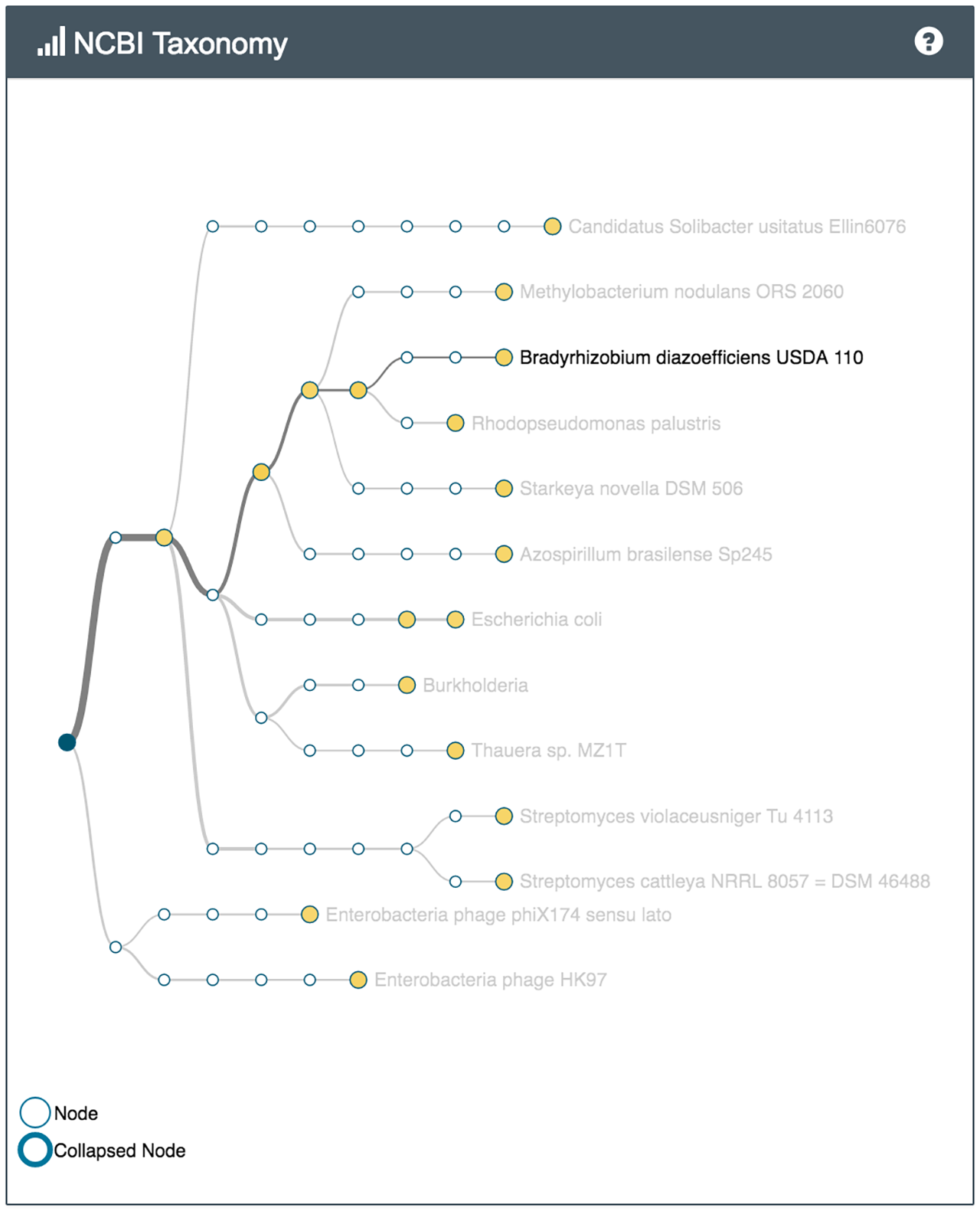
What's in my pot showing some of the species identified in the park sample (left) and the vegetable patch sample (right)
Cautious optimism
It is important not to over-analyse what we saw - the low read count meant that anything we didn’t see is as likely to have not been sequenced as absent in the sample, and anything we did see could easily be misinterpreted by the software as the wrong species. Also these were the results of just two samples. The entire experiment would need to be replicated in order to begin to make any kind of solid conclusions about inferring soil properties from their metagenomes using this method.

Watching our data come in on the MinION desktop software MinKNOW
Nevertheless the experiment can give broad encouragement that it should be possible to gain a rich understanding of a soil metagenome, and from this information infer properties of soil health and environment using MinION and WIMP. A distinct advantage of using MinION is that, with good quality data, you can make estimates about the proportions of different species in a sample, as this is related to the number of reads for each species.
Although there are still many technical challenges with using MinION, with lab space, some patience and guidance from someone with experience, it is a technology with remarkable potential. While it would take a lot of effort and significantly better microbial knowledge than we currently possess, the possibility of using nanopore sequencing to gain a better understanding of soil health is genuinely exciting.
If you are interested in discussing this project, our results or having a look at the data we collected, please get in touch!