Last month we travelled the world to discuss our BioDesign team’s work. We presented at the Pistoia Alliance UXLS Conference in Boston and the OpenVis Conf 2018 workshops in Paris.
We spoke about 5 principles of good information experience that we’ve found to be especially helpful in the design of software for the life sciences. In this post we explain these principles, why we think they matter, and how to use them in practice.
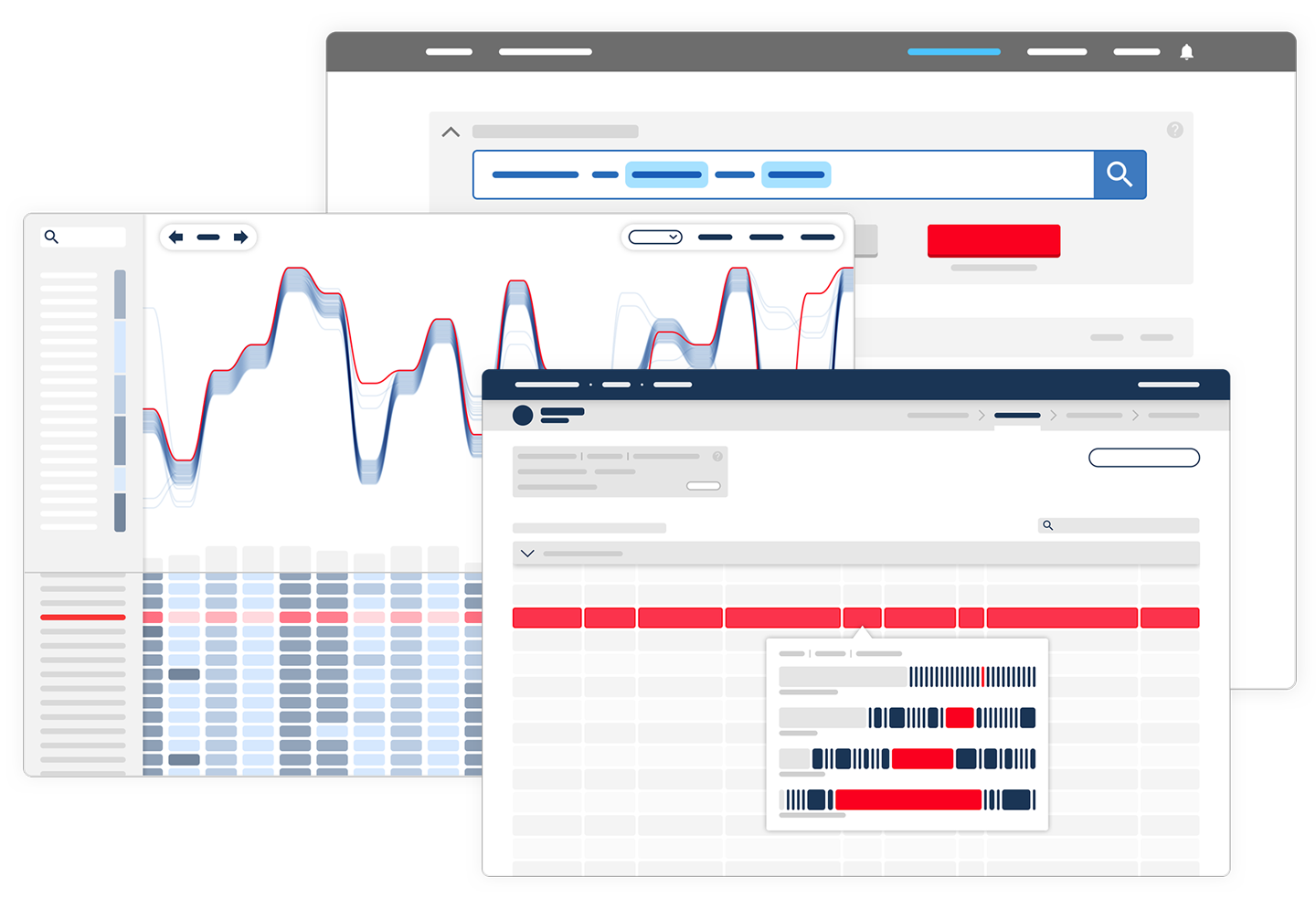
A key part of software design for the life sciences is understanding and shaping information experience. This means working to ensure that users know how data is processed, and offering opportunities to manipulate the ways in which a tool works. Most importantly it is about making it is as easy as possible to find patterns and insight in data.
Below we discuss 5 design principles that we use when designing software for the life sciences and how they can be implemented.
Transparency in Information Analysis
One of the main reasons for developing life science software is to automate some part of an analytical process through the use of algorithms, easing the burden on scientists. Often, most of the logic of an algorithm tends to be abstracted away from the users - it’s a ‘black box’ that no one can see into to make sense of.
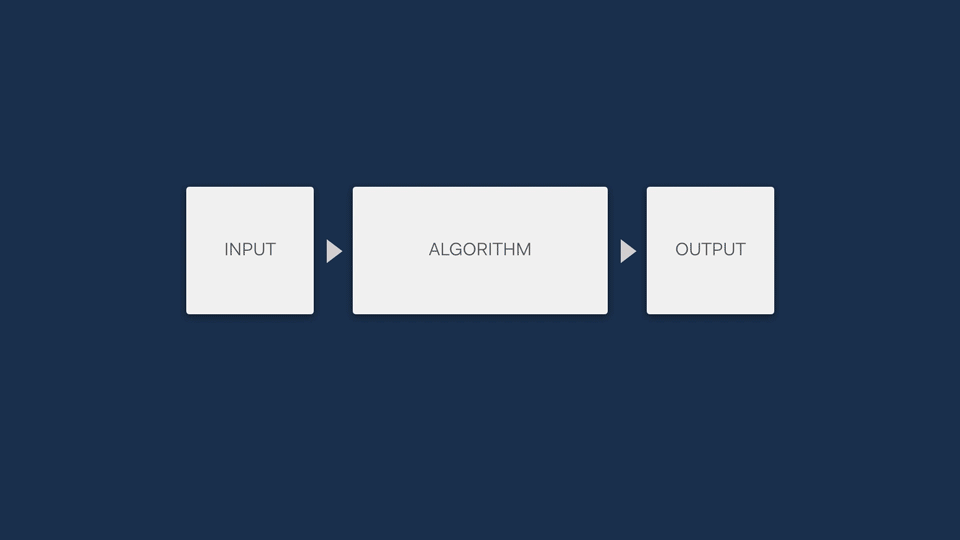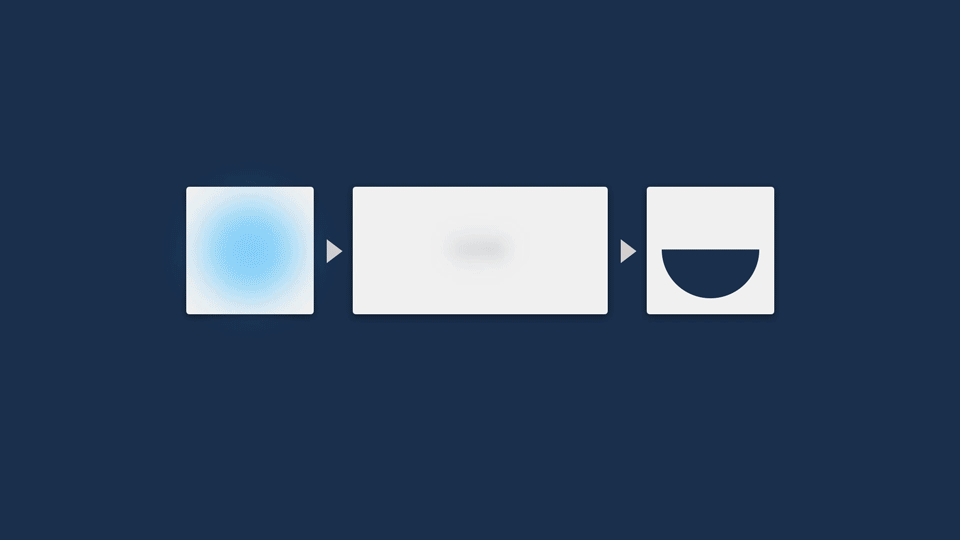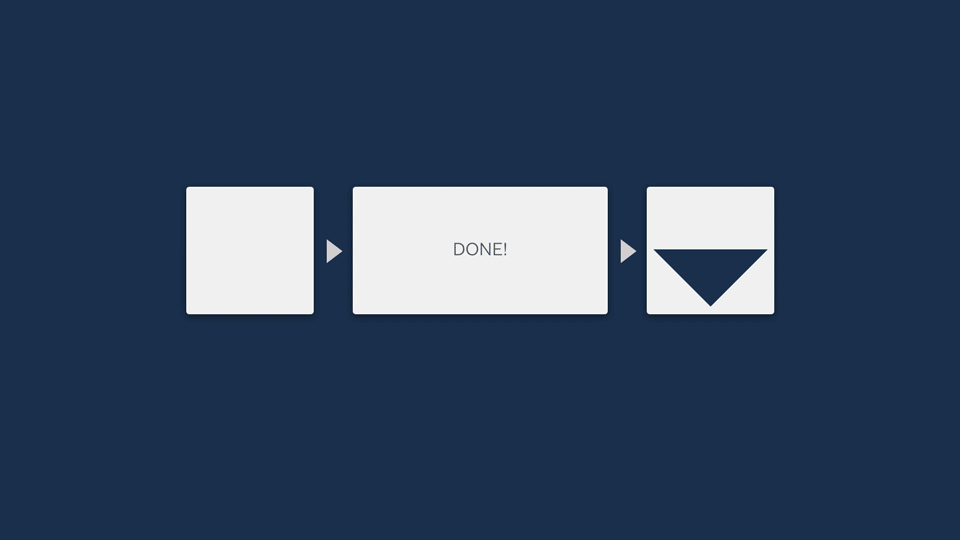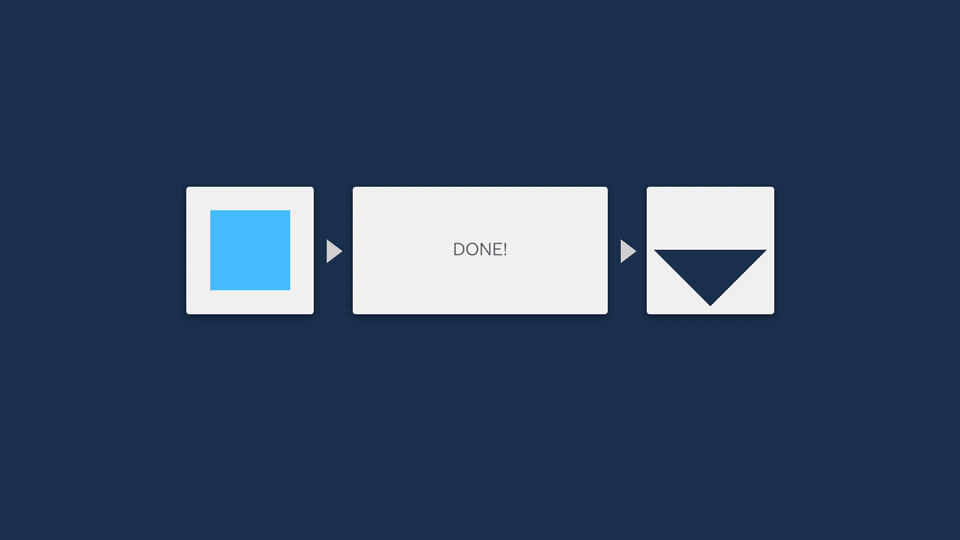
Unless people understand how a software algorithm works, they are unlikely to be able to trust it. For example, in clinical genetics labs like the one that Simon worked in, the sensitive nature of the work means that a tool won’t be used at all if lab scientists don’t have an understanding of the principles it is based on.
This means it’s a really good idea to find a way to open up the black box, showing the steps that an algorithm is making to allow users compare the mechanism of an algorithm to their own understanding of the process.
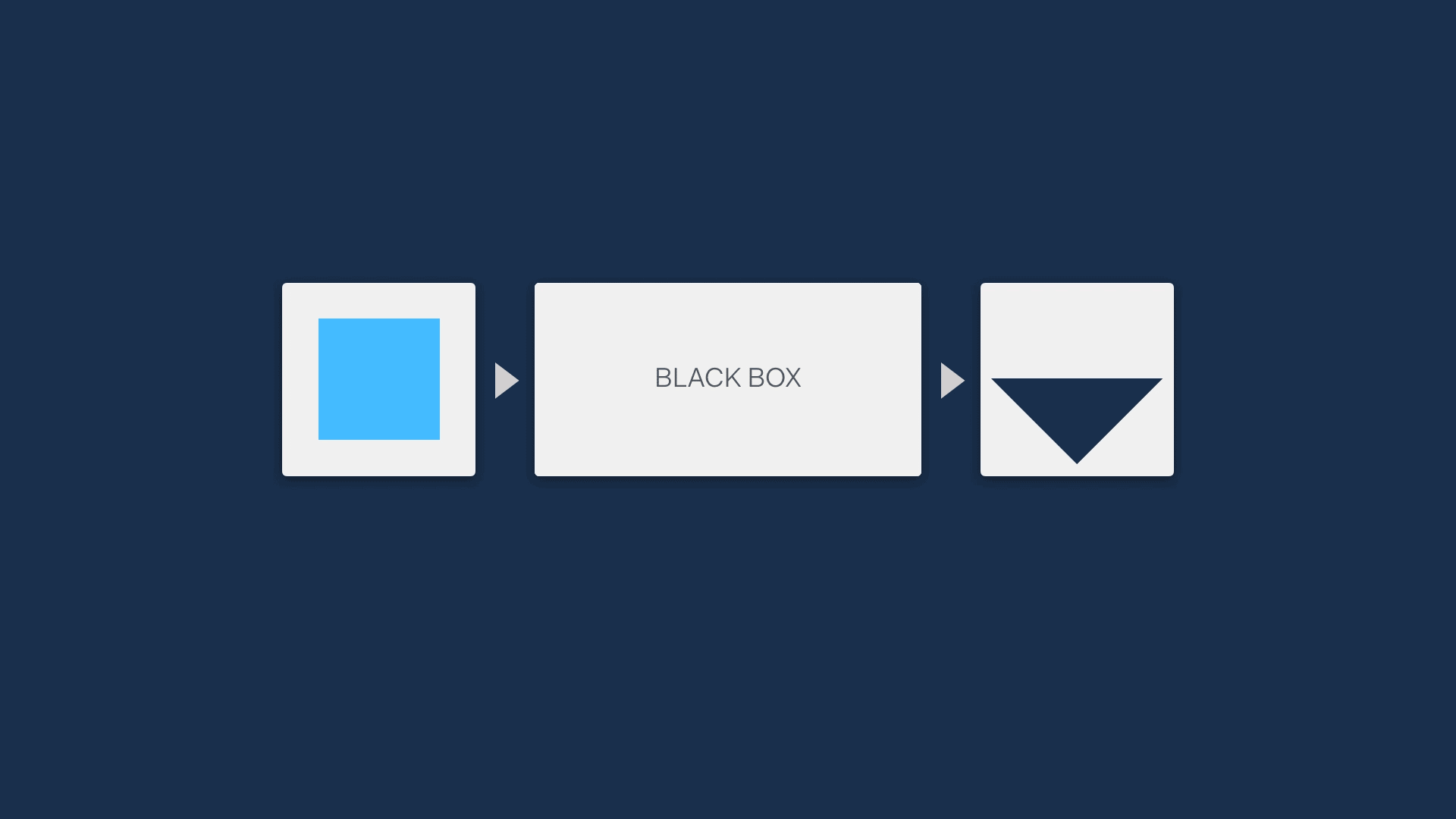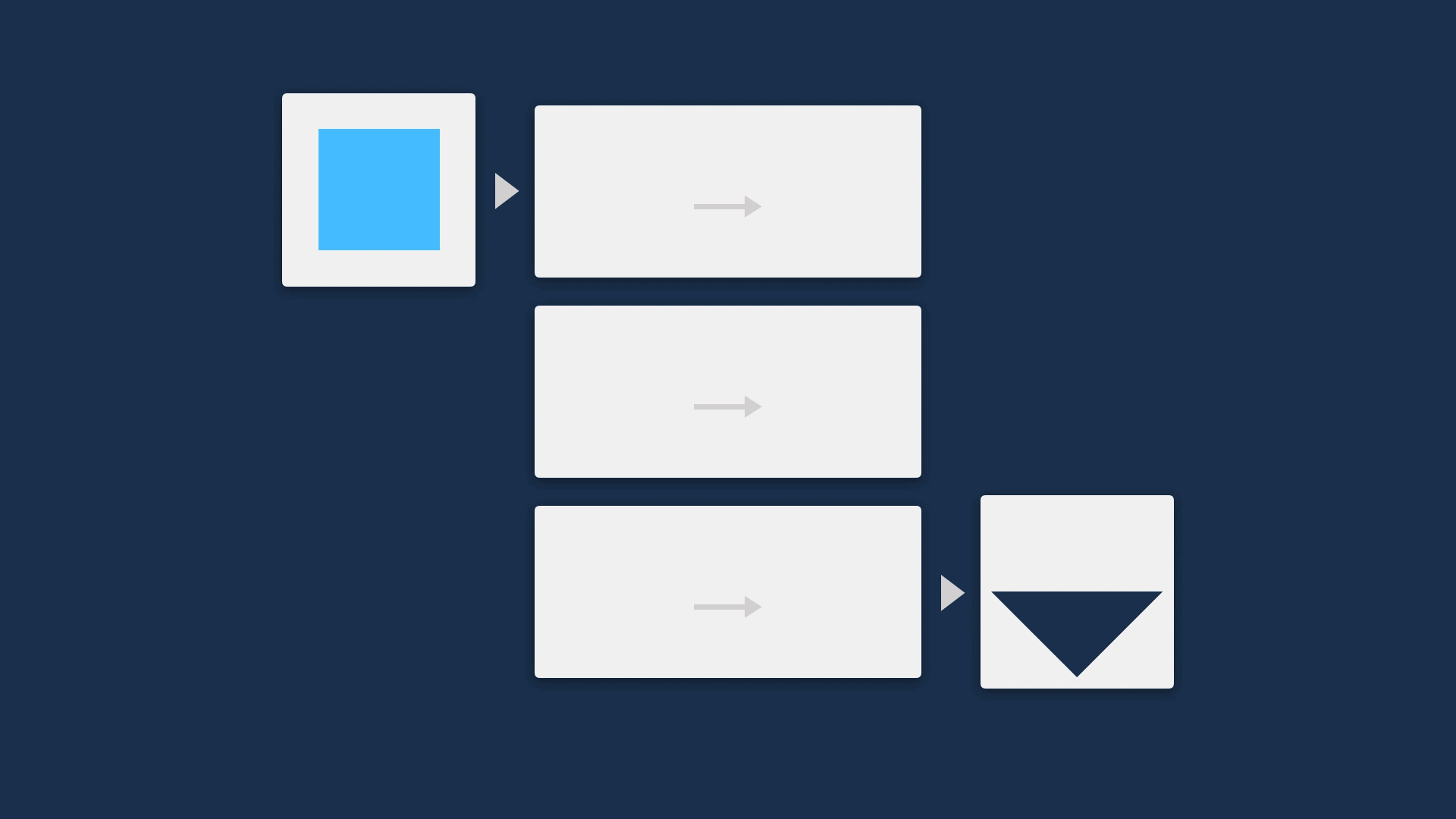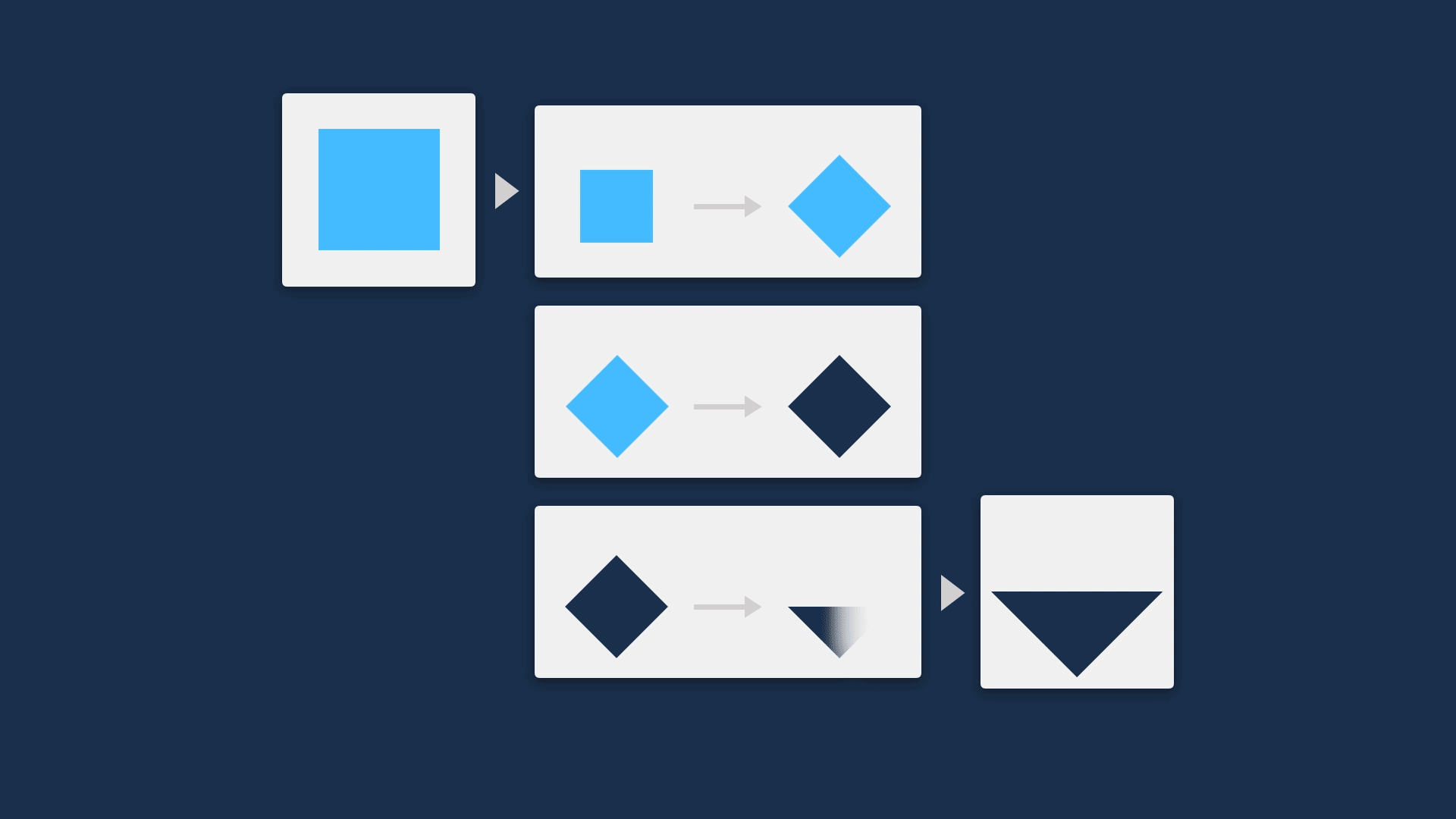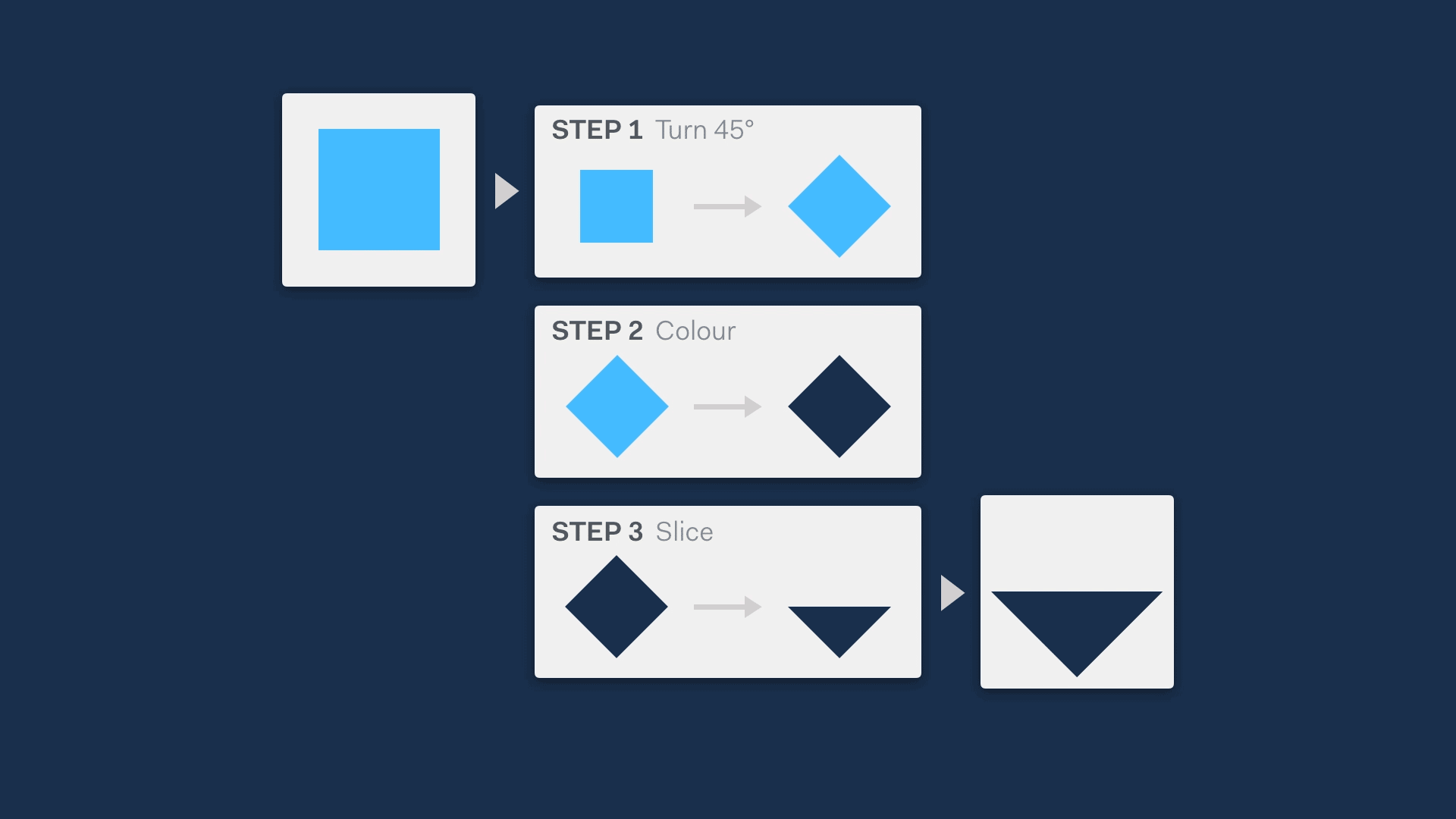
A great way to do this is visually. This can be a very quick method to describe functionality, while not revealing so much that IP is at risk.
Going further, if the method of revealing the steps in the process is also interactive, this not only increases the ease of understanding, but also makes it customisable — potentially increasing the utility and trustworthiness of the tool so it can be implemented in more contexts.
In our ‘Opening the black box’ case study we explain how we applied this first principle to help users understand the working of a clinical genetic analysis tool.
Information Hierarchy
A major challenge in the life sciences is the scale and richness of biological data. When using software tools for accessing or manipulating large biological datasets, it is easy to become overwhelmed, miss what you are looking for, and miss opportunities for discovery.
Understanding the information priorities of users – which data they want to compare and consume, and in what order – is key to preventing confusion.
In practical terms, applying the principles of information hierarchy can be quite simple. For example, this could be a matter of prioritising information using colour, size, and order of different information types.

Other approaches involve moving less important (or more technical) information a click away behind drop-downs or on secondary pages. Standard design patterns proven in other domains are equally useful in the life sciences.

It’s important to bear in mind that information priority is not universal. Depending on what they’re using a software tool for, users will have different opinions on what they need to see first, what second, and so on. They will also have preferences regarding the level of control they need over the system’s functionality.

For example, bioinformaticians may need to know the type of sequencing platforms and the exact pipeline settings used in their experiments, while clinical scientists may prioritise access to journal articles in which disease symptoms are discussed. This means that designing the right level of interface customisability for different user types relies heavily on user research to understand preferences.
Flexible Workflows
Scientific workflows can switch rapidly from the ordinary to the novel as researchers respond to signals in their data. Software interfaces need to support streamlined completion of routine tasks as well as facilitating detours for more in-depth data exploration.
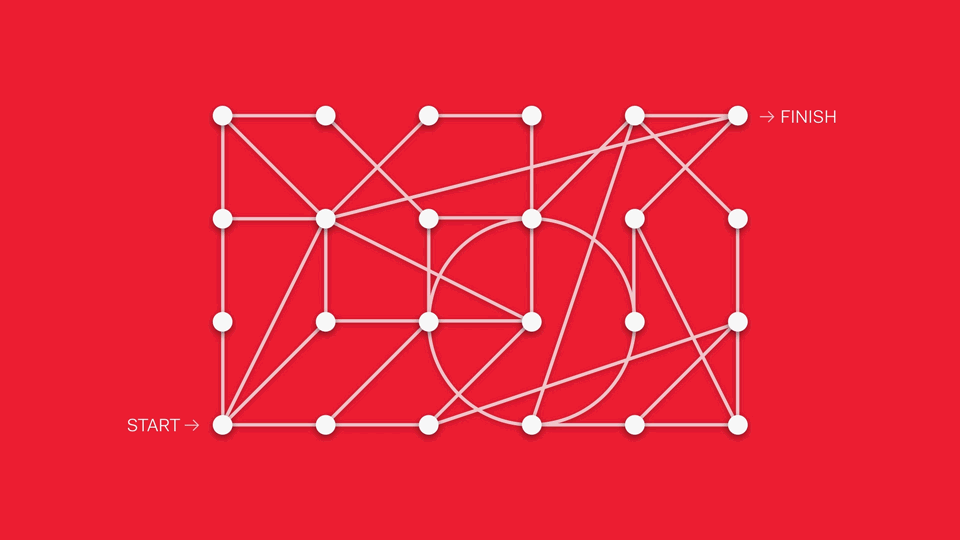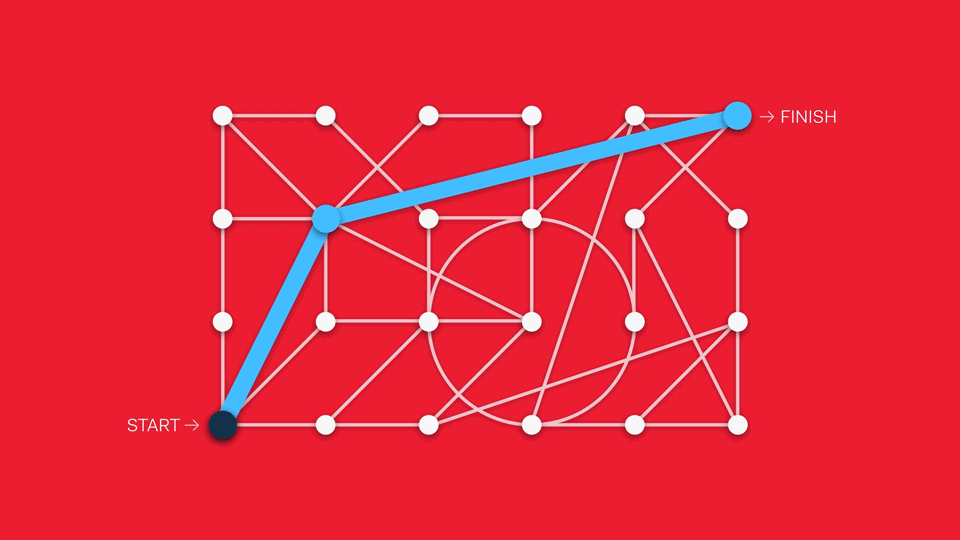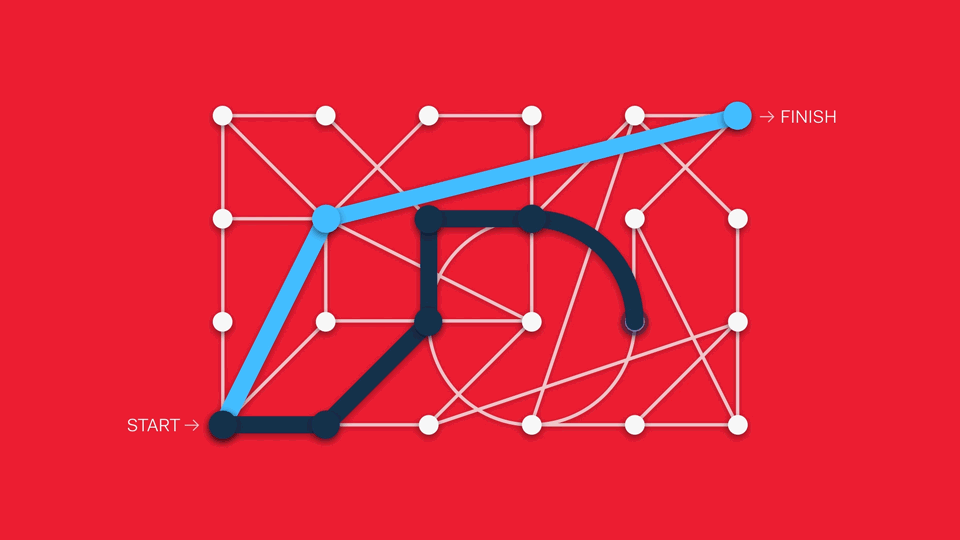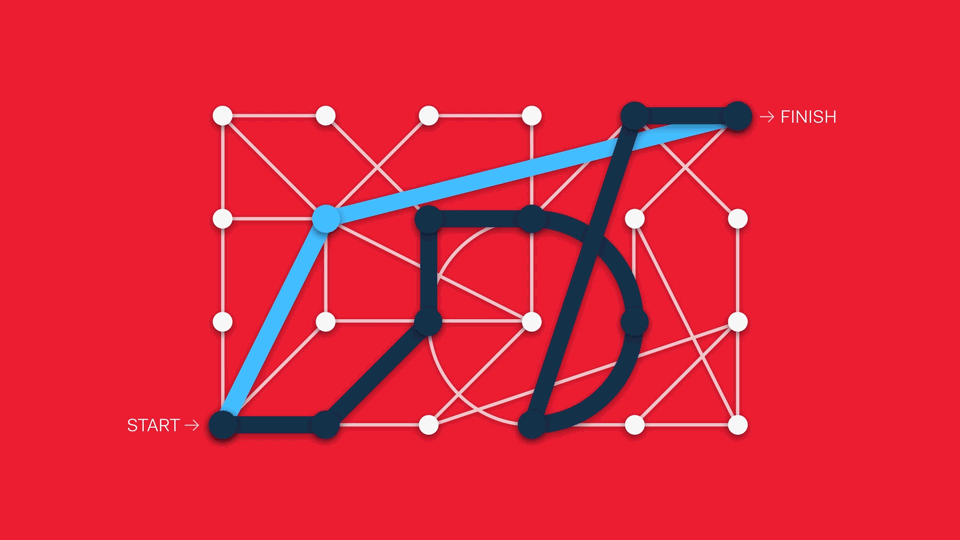
This can be challenging to implement in the life science context. Workflows can include multiple steps, protocols can vary from one lab to another, and best practices constantly evolve.
One way of addressing this is through treating different analytical features modularly, suggesting popular next steps from one module to another. Another approach is to pursue a sandpit style of software, in which there are no enforced workflows in favour of maximum flexibility (although this can create barriers to newcomers if the interface is too overwhelming).
Understanding what scientific users want to accomplish, their context, and their limitations is especially important for designing workflows that are optimal. At BioDesign we do this by observing existing patterns with currently used tools, and by prototyping and testing scenarios based on known use cases.
Encouraging Exploration of Results
Scientific information inherently lends itself to visualisation. Modern web browsers allow for visual representations that are multidimensional, dynamic, and interactive. However, because data in the life sciences is multidimensional and vast in size, it often makes it challenging to capture all significant information in a single visualisation format.
We believe it is important to give people the opportunity to view their data from a variety of perspectives, making it possible to find patterns, pull insights out of data, and generate new hypotheses.
Of course, each visualisation type has its own strengths and weaknesses. Each graphic will highlight or emphasise certain aspects of the data, but may obscure or distort others. By paying close attention to what these factors are, and testing extensively with users, it is possible to develop novel and complementary visualisations that improve a scientist’s ability to making a discovery.
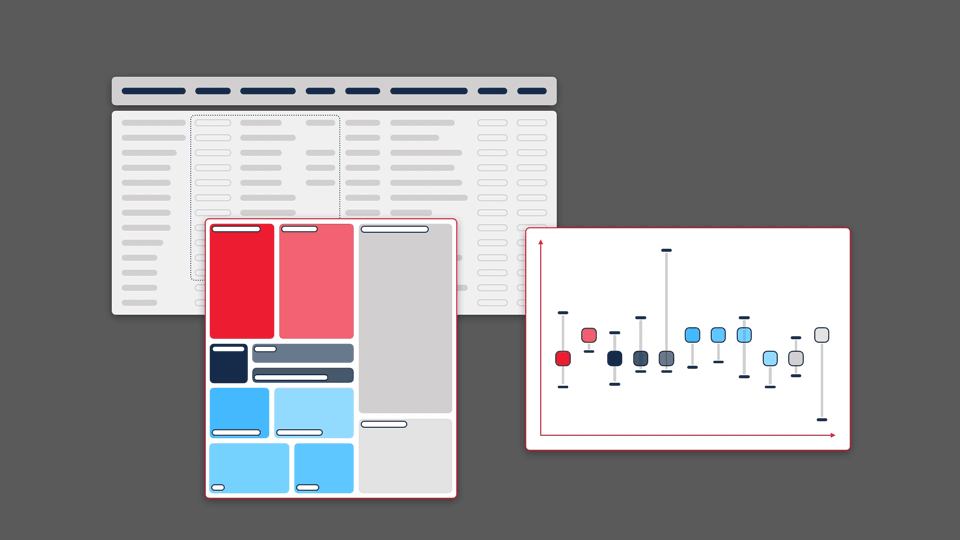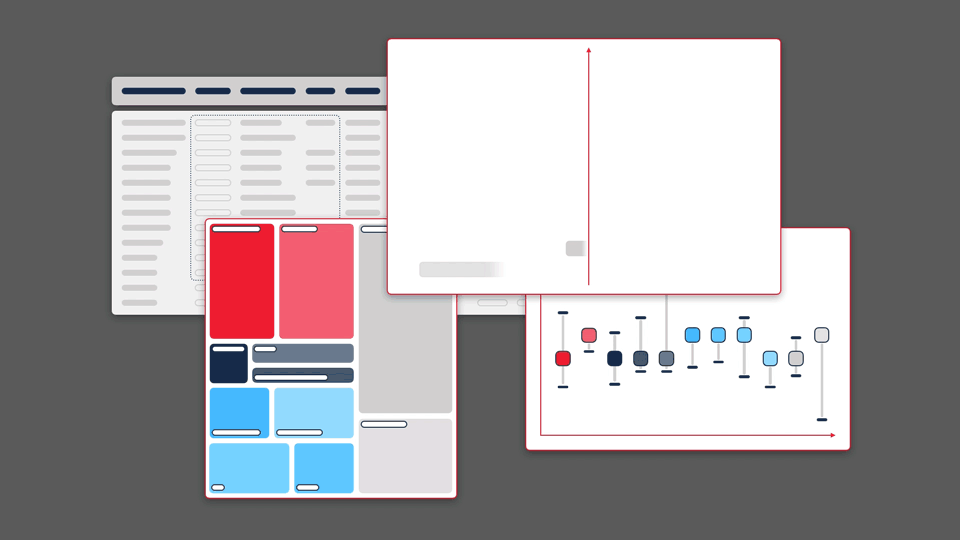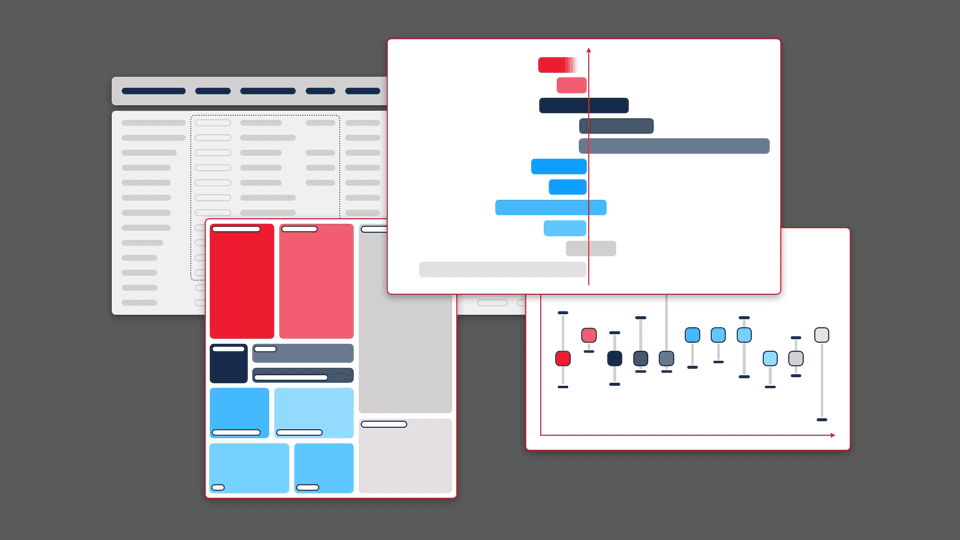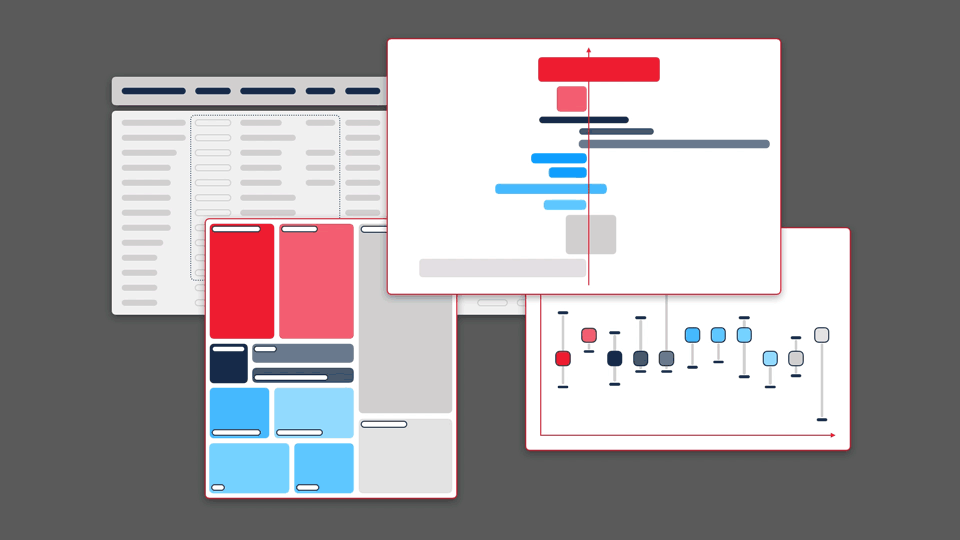
This principle guided our work for Sequence Bundles — a novel method for visualising sequence alignments that we designed and published as an open-source software tool. Sequence Bundles users can expose protein and DNA patterns that other visualisation methods would otherwise fail to surface.
Design-led Research
User research is essential to producing good software. Without it, it becomes easy to over-engineer functionality, or build confusing user interfaces (UIs). It is especially important for software development in the life sciences, where functionality is often complex and there are fewer instances of known design patterns that can be reliably followed.
At BioDesign, we use a design-led research approach in our work. This means that we produce design prototypes as early as possible in which we capture our best understanding of user interactions, workflows, information hierarchies, and data visualisations. We then use these prototypes in research interviews to test ideas and assumptions with user and experts.
A prototype can be a click-through mock-up, a diagram of a workflow, a screen from a proposed software UI, or a map of ideas… Anything that clearly indicates the concept or proposal that you want to test with users. Interviews should be recorded, then insights are pulled out and collated to build up a picture of a user’s needs, understanding, and intentions. Using this feedback as a basis for iteration allows for rapid improvement and refinement in the design of software.
We have found the design-led research approach to be most effective in the early stages of development, where open user feedback is instrumental in defining features, interaction models, and the scope of life science software.
Design Matters
Many hopes are placed in modern life science software: from providing genetic diagnoses to patients, to automating complex experiments, to identifying new drug targets, to organising entire domains of knowledge. Pivotal to all these promises is how we interact with information and data. Designing good information experiences for the life sciences will help these tools to meet their potential.
At the most basic they will be more efficient, but they can also be more compelling, delightful, and understandable. At their best, well-designed information experiences will enable scientists to find patterns that would otherwise have been missed, and formulate new hypotheses that can push research forward.