Running our own scientific experiments is a really important part of our work at Science Practice. However this isn’t always straightforward, not least because we don’t have our own lab. To get around this problem, recently we’ve been using a system we’re calling our ‘distributed lab’ - taking advantage of the networks and spaces available in London to get our experiments done.
Our most recent experiment has been in genetics. We’ve been trying to find out if we can use a new sequencing technology, called MinION, to identify different microorganisms found in soil using their DNA. Our idea was to see if it’s possible to use this data to understand soil health, as well as testing if we could independently run our own sequencing experiments.
Our findings from the experiment are discussed in a separate blog post, here we focus on our experience and learning from setting up our first sequencing experiment.
Our experiment
Up until very recently our goal of independently running genetic sequencing experiments would not have been possible; the cost of equipment was simply too high. However the introduction of Oxford Nanopore’s genetic sequencing device MinION has changed that. This new tool reads genetic code in a very different way to other available sequencing technologies, reducing the preparation cost and time for samples as well as allowing the device to be an order of magnitude smaller than other pieces of tech, making it portable.
Oxford Nanopore’s ultimate company aim is:
To enable the analysis of any living thing, by any person, in any environment.
This is a concept we find enormously exciting, adding to our desire to test out how easy it is for companies and people like us to organise experiments without a dedicated lab space.
Getting started
Before we went off on our own, we decided to get some help from people with experience in using MinION. We contacted researchers at the University of York as well as Public Health England who generously agreed to let us run trials with them. While the MinION protocol is relatively straightforward, the opportunity to learn from people with expertise was incredibly valuable. We’ve written about our experiences in these trials here and here.


Our trial experiments at Public Health England (left) and York (right)
Going out on our own
After our two trial runs we felt we were ready to run the MinION protocol independently. However before we could do this we needed to find a space in which to do it.
Initially we considered buying equipment and consumables (such as pipettes and ultra pure water) to run the MinION in our offices. However after the trials, it became clear that while it would be possible to run MinION in an office, it would be too costly for a proof of principle experiment.
Finding Space

The London BioHackspace
After deciding not to use our office as a lab, we needed to find an alternative space in which to conduct our MinION Sequencing experiments. Previously we had success working in academic labs, and of course had already made contact with researchers who may be willing to let us continue our experiments. However for this project we wanted to understand the demands of running sequencing experiments independently, and so decided against going down this route.
A big factor in this choice was our discovery of the London BioHackspace. This is a community-run lab in London that provides access to basic experimental equipment for a small monthly fee. Anyone is able to use it, provided they are given an induction and adhere to safety rules. The lab is built on a model of sharing equipment and knowledge between members, and makes perfect sense for anyone who is looking to find proof of concept of a practical scientific idea.
While we had found a space, and the bulk of the equipment needed, there were a few pieces of specialised equipment and reagents that weren’t (yet!) available at the BioHackspace.
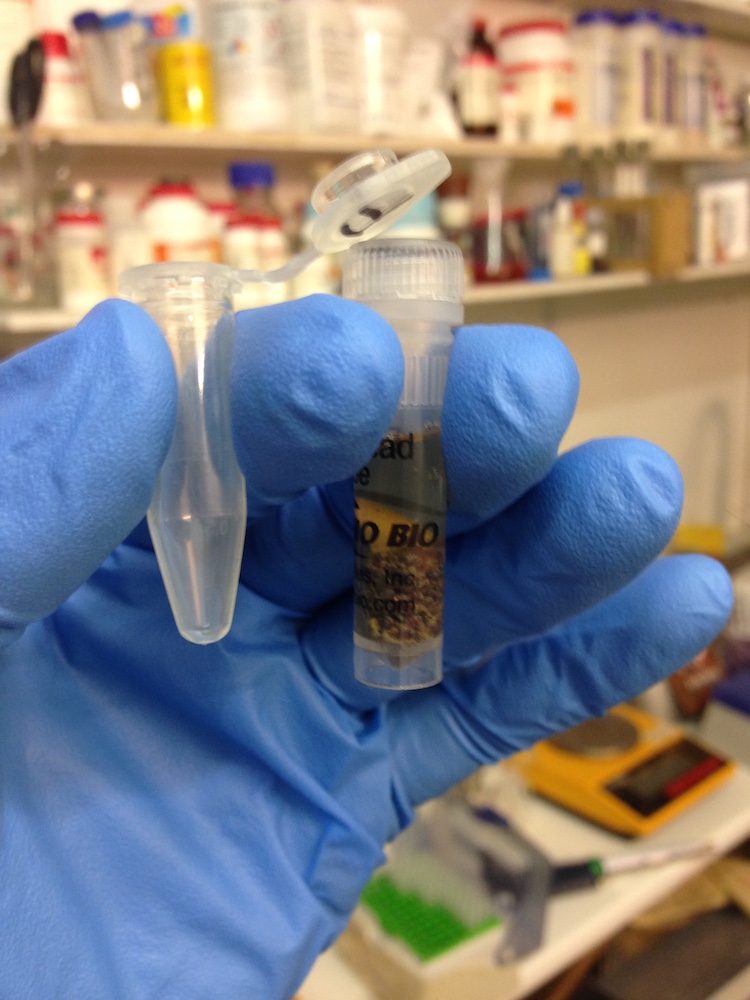
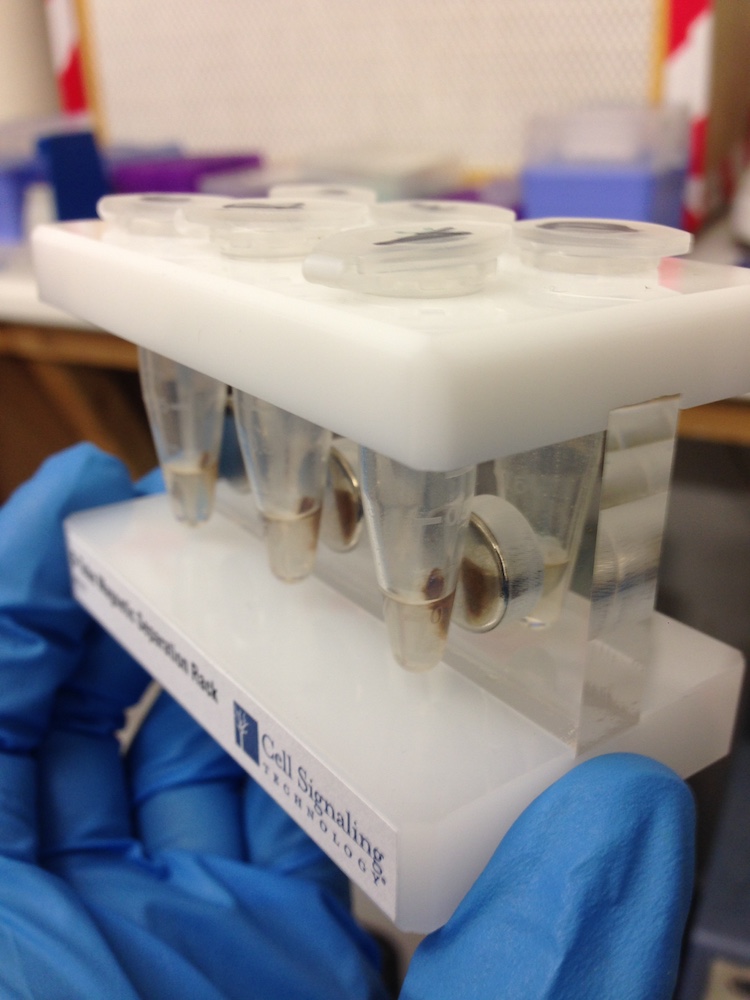
DNA extraction kit (left) and DNA attached to brown magnetic beads in solution.(right)
These missing components presented varying levels of difficulty for us to get hold of. Some things we were able to purchase such as DNA extraction kits to isolate DNA from soil, or DNA binding magnetic beads and a magnetic sample tube rack that enable separation of DNA from liquids.
One piece of equipment that was a particular hurdle was a Qubit fluorometer. This device is able to measure the concentration of DNA very precisely, which is essential information when setting up a MinION sequencing run, but is prohibitively expensive for single MinION experiments.
After some research we discovered ‘iLabs’. This is a system that allows people to book equipment in professional service labs. Ordinarily this is intended for use for people in academic labs, giving them the option of hiring equipment rather than buying outright. However, after discussing our project with the Genome Centre at Queen Mary University they were happy to let us book time on their Qubit.

The Genome Centre at Queen Mary University
Another important component of organising experiments is the ordering of reagents. Essentially, this requires setting up accounts with suppliers of biological experimental reagents so that they will allow you to purchase from them. In our experience none of the suppliers we attempted to order from had any issue with this, although we were asked to explain our intentions very clearly. The process was likely made simpler as we weren’t attempting to purchase anything that carries any significant health risk.
Overall, the process of running our first independent genetic sequencing experiment provided us with some useful lessons. While individually none of the organisational steps were particularly complex, they often took longer than we expected. For example, as we are not an academic institution, we weren’t able to fill out applications to buy reagents as suppliers expected. This ultimately wasn’t an issue, but it did take a couple of days of back and forth emails to resolve, pushing back the experiment.
What we did find is that people are often very generous with their time and expertise. By contacting people directly and explaining your goals and motivations, it is often possible to organise a way to get what you need for your experiment. While not every city has a community lab space like the BioHackspace, making contact with academic researchers at a local university can be a viable alternative.

The MinION running at the BioHackspace
It is important to note that running experiments this way can be somewhat risky, as the learning curve when trialling new scientific experiments can be steep, with few safety nets for mistakes. It is also time consuming. For us the distance between our office, the Hackspace and the Genome Centre wasn’t excessive, but was definitely not the easiest way to work. In particular, pausing experiments to allow for sample transportation has to be well planned, to minimise impact on results.
Distributed Labs
The idea to enable anyone with an interest in conducting their own scientific experiments to pursue that interest is something that has gained popularity in recent years through the citizen science movement. However, citizen science commonly focuses on more everyday equipment such as wearable technology and smartphones. Experiments requiring a laboratory stocked with pipettes and centrifuges, are more unusual, placing different demands on the organisers.
The process of organising space and time to conduct MinION sequencing experiments has highlighted to us the amazing potential of using community spaces and hired time in service labs. This ‘distributed laboratory’ model has enabled us to conduct our own proof of concept experiments using cutting edge sequencing technology. This is something that we were not certain was possible before we began the project.
The opportunity to run our own genetic sequencing experiments, and gathering first hand experience of the whole process of sample collection to data analysis was incredibly exciting, informative, and ultimately rewarding.
Checklist for using Distributed Labs.
-
If you are interested in trialling something that you’ve not done before, try asking researchers with experience in that field. Not all have time to help, but many are enthusiastic to share their knowledge with outsiders.
-
Try and find out if there is a community lab near you, and become a member. If not, try contacting researchers interested in a similar topic. There will be a limit to the time and resources they can spare, so you may have to tweak your project to be inline with their interests.
-
Alternatively you could set up your own community lab. This is a big undertaking, but very valuable. For help and advice try here, or contacting the BioHackspace.
-
Service labs sometimes allow people to hire time on specialised equipment. Email them, explain what you want to do and you’ve got a good chance at access.
-
Provided they are not harmful, reagent suppliers are likely to allow you to order from them. For molecular biology we used Thermo Fisher, Beckman Coulter, New England Biolabs and Qiagen.
-
MinION and Oxford Nanopore have an active and knowledgeable customer support, as well as an engaged community of people using their devices. If you need help, ask!
If you would like to talk to us about our experiences in setting up a distributed lab, please get in touch!